Real_data_filter
Margarita Orlova
January 28, 2020
Last updated: 2020-02-05
Checks: 6 1
Knit directory: Thesis_single_RNA/
This reproducible R Markdown analysis was created with workflowr (version 1.5.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191113) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| C:/Users/Moonkin/Documents/GitHub/Thesis_single_RNA/analysis/MLE.py | analysis/MLE.py |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Unstaged changes:
Modified: Data_sim.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
In this vignette we will load the data, pre-process it and analyse top gene by p_beta parameter.
Filtering
Here we load raw dataset and filter it by cell, individual parameters and order genes by smallest p beta parameter
library(dplyr)Warning: package 'dplyr' was built under R version 3.5.3
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionraw_data<-read.table("D:/Uchicago/Thesis/Real_data/scqtl-counts/scqtl-counts.txt", header = TRUE, sep = "", dec = ".")
combined_filter<-read.table("D:/Uchicago/Thesis/Real_data/combined_filter.txt", header = FALSE, sep = ",", dec = ".")
combined_filter[,1]<-combined_filter[,1]+2 #adjusting index from python to R
data<-data.frame(raw_data[,1],raw_data[,combined_filter[,1]])
colnames(data)[1]<-"gene"
mean_txt_file<-read.table("D:/Uchicago/Thesis/Real_data/mean/mean.txt", header = TRUE, sep = "", dec = ".",row.names=NULL)
gene_filter<-mean_txt_file[,c("gene","p_beta")]
data<- left_join(data, gene_filter, by = "gene")Warning: Column `gene` joining factors with different levels, coercing to
character vectordata <- data[order(data$p_beta),]
clean_data <- data[,1:(ncol(data)-1)]Maximum likelihood estimation for one gene
Below we take the gene with the smallest p beta and estimate the parameters for all individuals that passed quality control
library(reticulate)Warning: package 'reticulate' was built under R version 3.5.3library(robustbase)Warning: package 'robustbase' was built under R version 3.5.1source_python("C:/Users/Moonkin/Documents/GitHub/Thesis_single_RNA/analysis/MLE.py")estimate_MLE_one_gene<-function(full_data){
data_ind<-substr(colnames(full_data),0,7)
data_ind<-data_ind[2:length(data_ind)]
est=matrix(, nrow = (length(unique(data_ind))-1), ncol = 3)
info=matrix(, nrow = (length(unique(data_ind))-1), ncol = 2)
iter=0
for (i in unique(data_ind)){
if (i=="NA18498"){
next}
iter=iter+1
x=full_data[ , grepl(i, names(full_data))]
x=t(data.matrix(x))
est[iter,]=MaximumLikelihood(x)
info[iter,1]=i
info[iter,2]=dim(x)[1]}
return(list(est,info))}top_gene<-clean_data[1,]
x=estimate_MLE_one_gene(top_gene)estimates=x[[1]]
info=x[[2]]
par(mfrow=c(1,3))
hist(log(estimates[,1]), main="Distribution of k_on estimates", xlab="", ylab="Number of individuals")
hist(log(estimates[,2]), main="Distribution of k_off estimates", xlab="", ylab="Number of individuals")
hist(log(estimates[,3]), main="Distribution of k_r estimates", xlab="", ylab="Number of individuals")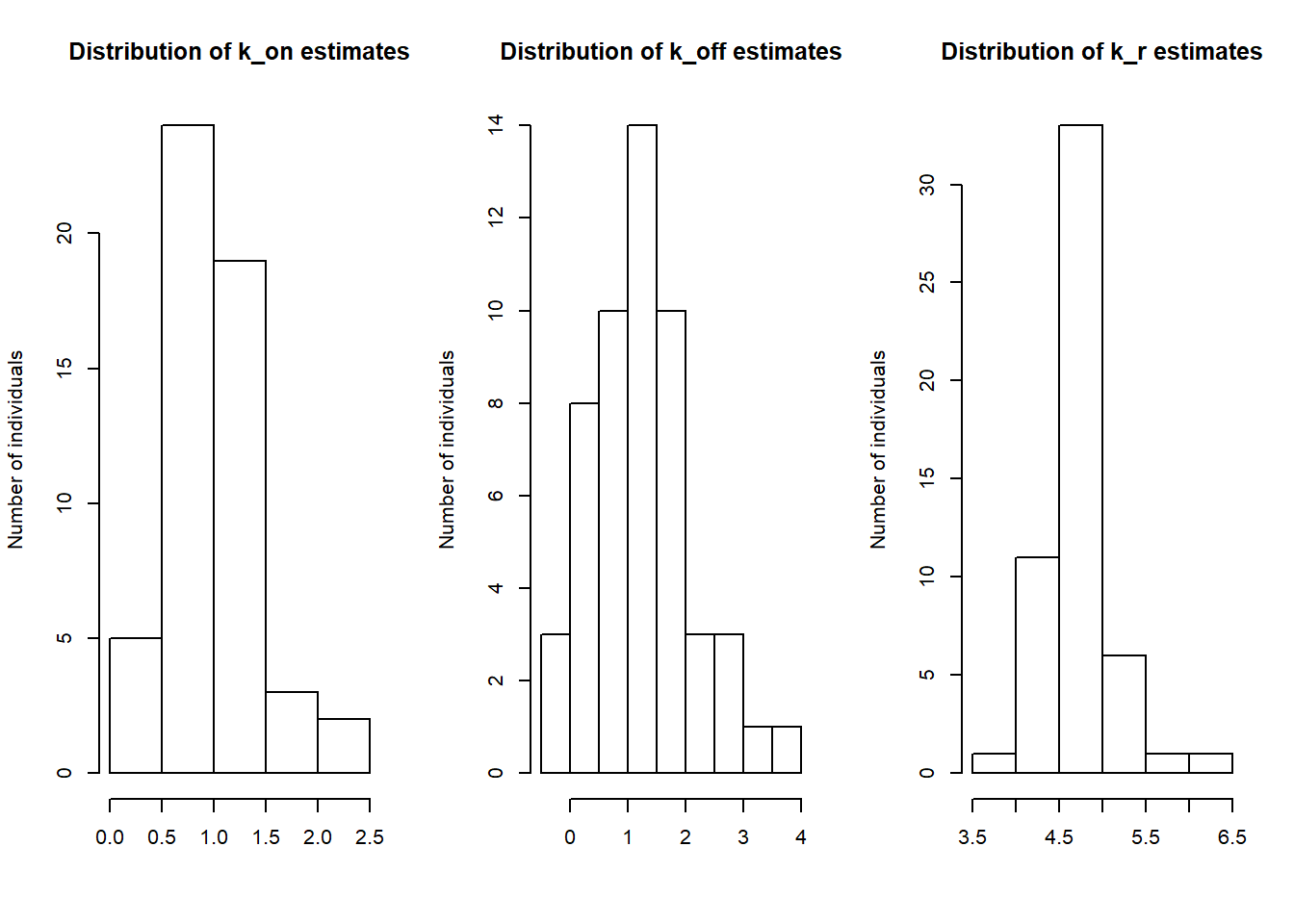
Distribution for one individuals
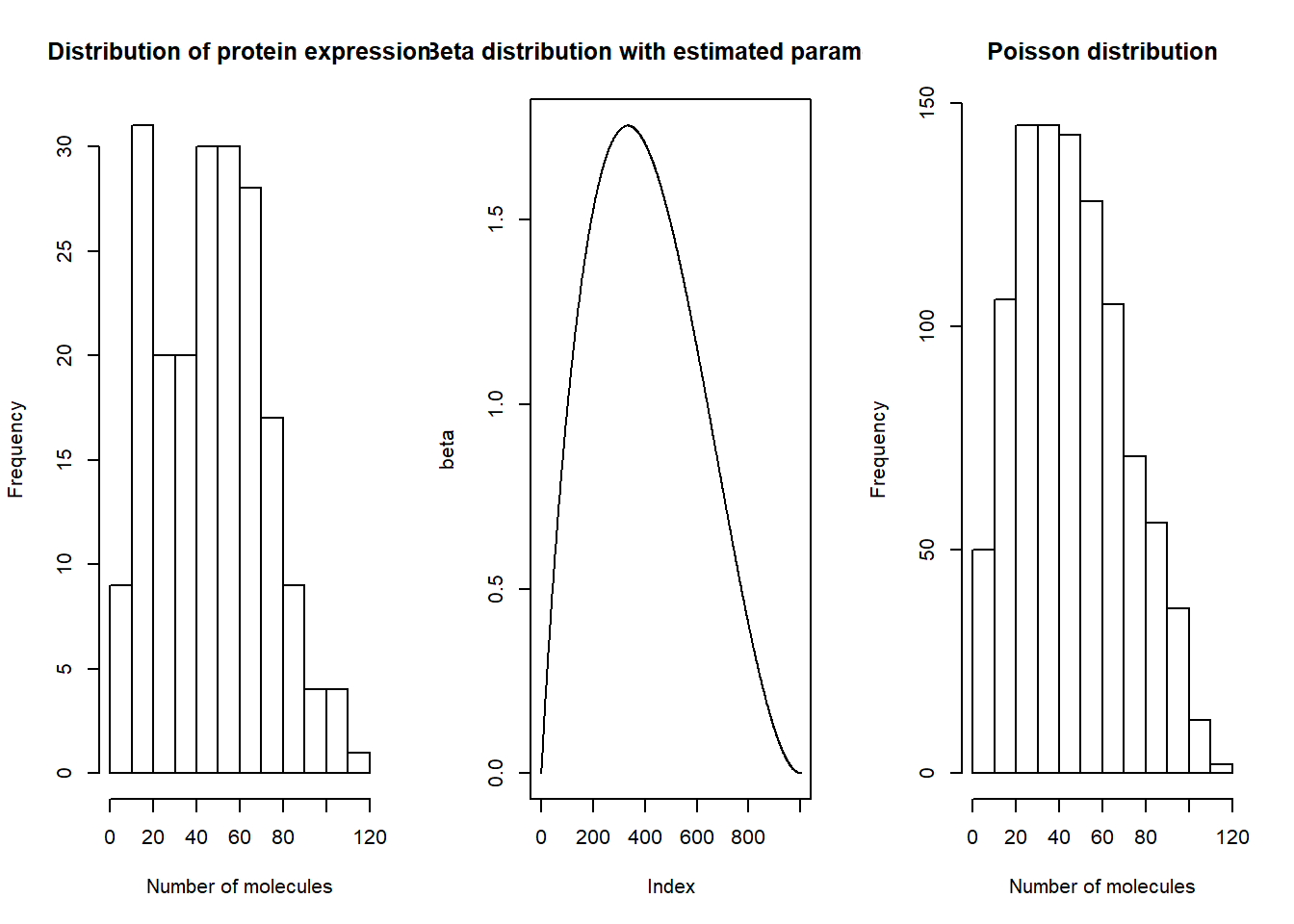
Then we take the individual with most cells (NA18501) and take a look at the estimated parameters:
The first plot is the real protein level distribution for NA18501 individual. Second and third plots are beta and poisson distributions with the estimated \(k_{on}\),\(k_{off}\) and \(k_r\) parameters.
Boxplots by genotype
We can get the genotype of the individuals and compare it to the estimated \(k_{on}\), \(k_{off}\) and \(k_{r}\) parameters.
Warning: Column `ind` joining factors with different levels, coercing to
character vector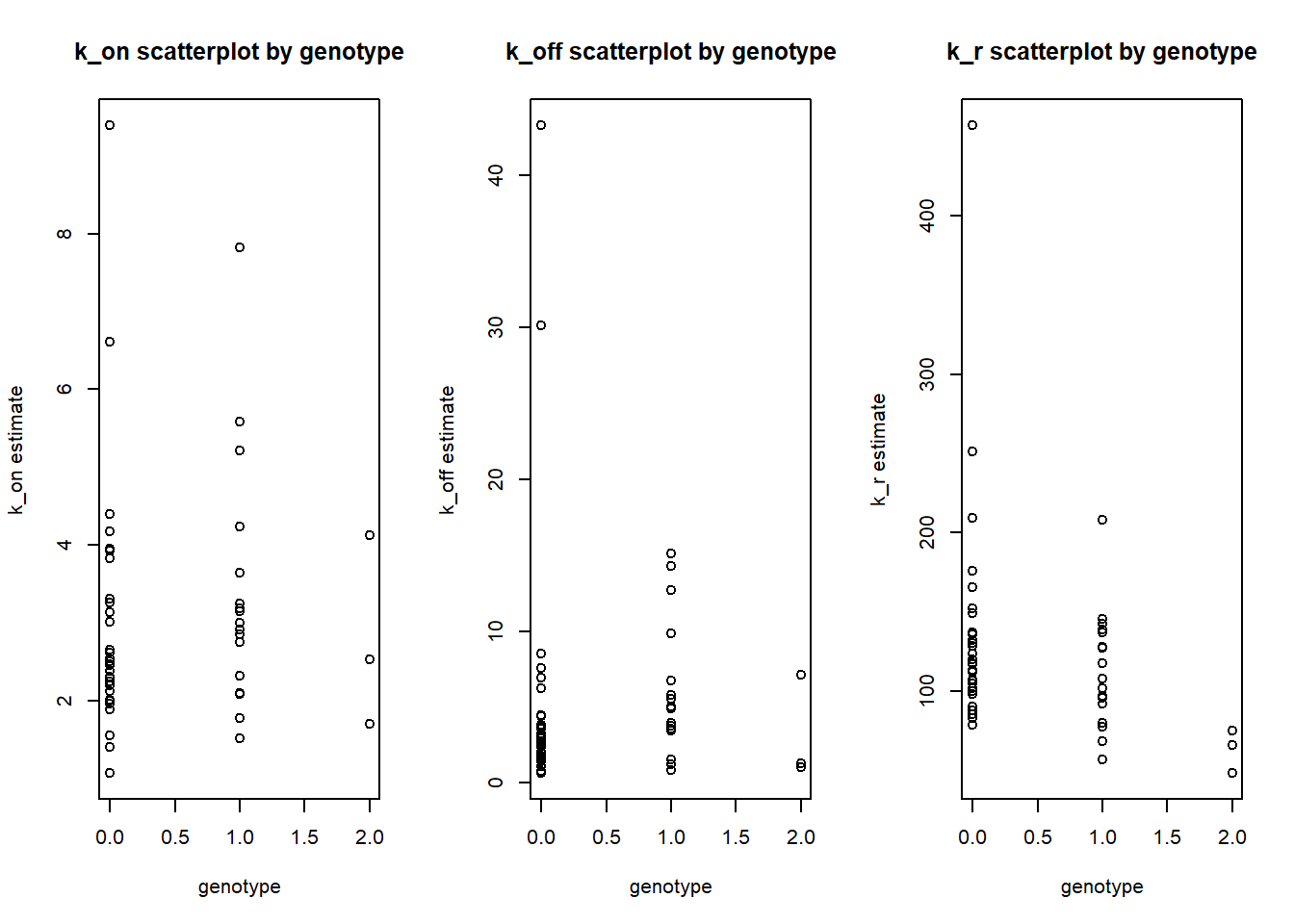
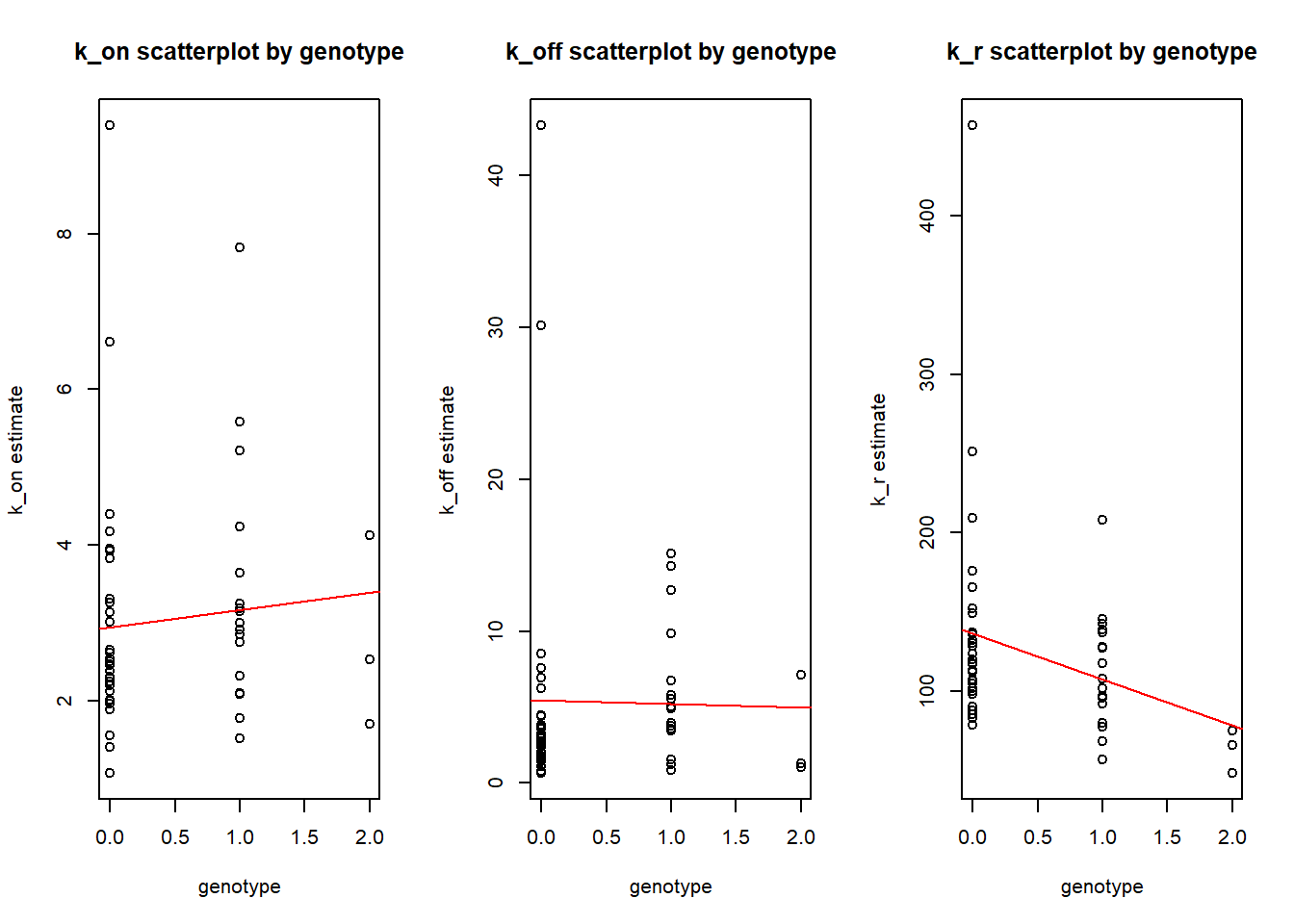
Regression
Finally we can fit the regression to estimate linear dependance of genotype on estimated parameters.
sessionInfo()R version 3.5.0 (2018-04-23)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 17763)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] robustbase_0.93-3 reticulate_1.13 dplyr_0.8.3
loaded via a namespace (and not attached):
[1] Rcpp_1.0.2 knitr_1.20 magrittr_1.5 workflowr_1.5.0
[5] tidyselect_0.2.5 lattice_0.20-38 R6_2.3.0 rlang_0.4.2
[9] stringr_1.3.1 highr_0.7 tools_3.5.0 grid_3.5.0
[13] git2r_0.26.1 htmltools_0.3.6 yaml_2.2.0 rprojroot_1.3-2
[17] digest_0.6.17 assertthat_0.2.0 tibble_2.1.3 crayon_1.3.4
[21] Matrix_1.2-14 purrr_0.2.5 later_0.8.0 fs_1.3.1
[25] promises_1.0.1 glue_1.3.0 evaluate_0.11 rmarkdown_1.10
[29] stringi_1.1.7 DEoptimR_1.0-8 pillar_1.4.2 compiler_3.5.0
[33] backports_1.1.2 jsonlite_1.5 httpuv_1.5.1 pkgconfig_2.0.2